The available modules of the GC-MS workflow can be grouped into three categories: pre-processing, statistics, and annotation (Figure 1). The modules for statistical analysis can also be used with processed data (sample x variable tables) coming from LC-MS, NMR or other "omic" technologies.
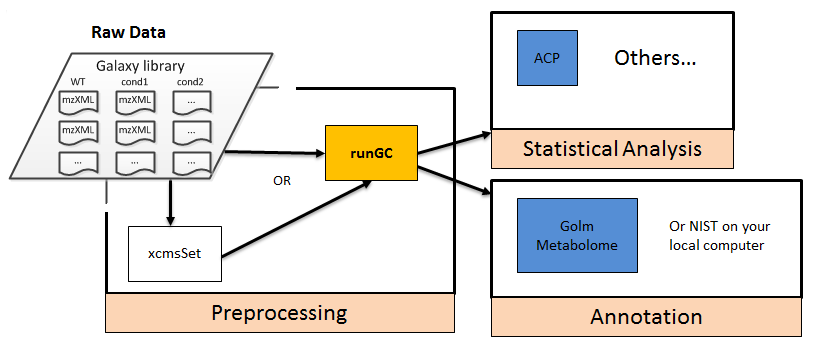
Figure 1: Available modules for GC-MS analysis. Raw data (in .mzXML or .cdf formats to be handled by metaMS::runGC) can be uploaded by using an FTP protocol or as Zip archive. Pre-processed data (i.e. variables x samples data matrix and the two associated tables for samples and variables metadata) can also be uploaded directly via the Galaxy interface.
Modules can be selected and chained by the user via the workflow editor. A dataset (named “GCMS_Idealg_FWS_SWS”') is provided as an example. It contains:
- 12 GC-MS spectra of algae samples (GC-MS simple Quad spectrometer)
- 4 replicats per conditions
- Condition 1: Fresh water strain in 1.6 ppt NaCl
- Condition 2: Fresh water strain in 32 ppt NaCl
- Condition 3: Sea water strain in 32 ppt NaCl