The community offers a large choice of public or private resources helpful in spectral annotation steps. Classical strategies are based on queries with accurate mass on large or specialized chemical compounds banks, dedicated metabolites and small molecules databases. Moreover, web services allow applications to communicate directly with all resources and propose a high speed service compatible with thousands of queries.
In W4M, we propose the integration of search tools for the following banks: the Human Metabolome DataBase, Lipidmaps, Kegg-compounds and for ChemSpider a search meta-engine.
To complete analysis practices, we provide a mass search tool using the spectra repository MassBank and a formula elucidator based on the HiRes [12] algorithm and implemented by the Fiehn laboratory.
As a conclusion, we provide on W4M 6 annotation tools, with the additional possibility to chain them or use each in parallel. The workflow mode of Galaxy allows experts to design and share annotation strategies (Figure 7).
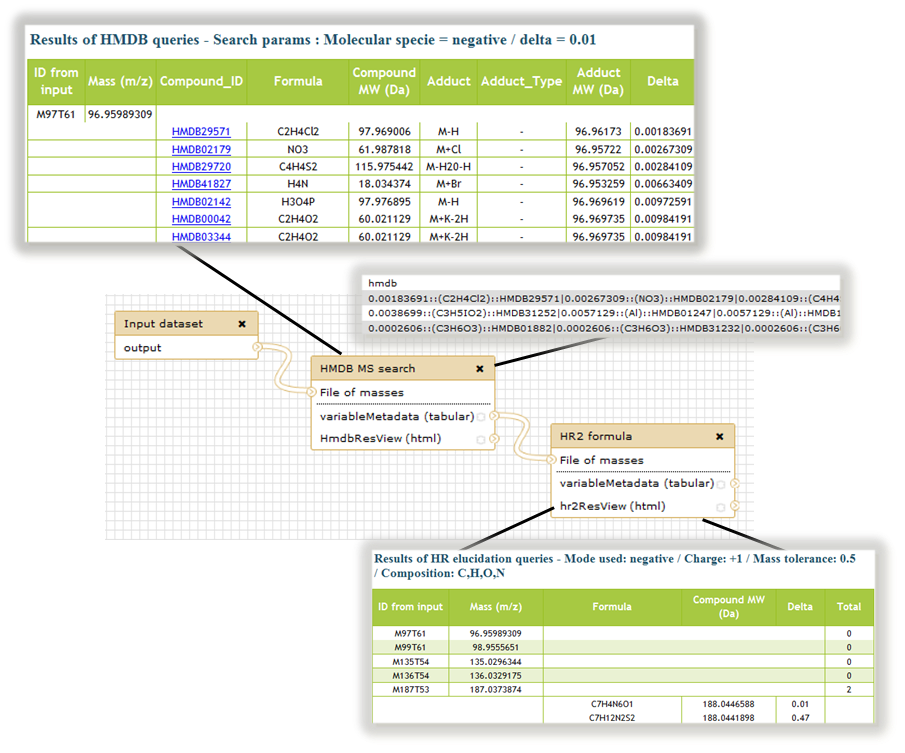
Figure 7 : Annotation sub-part of W4M: Workflow and outputs results views of a possible annotation strategy merging search on a reference bank (HMDB MS search tool) and in silico formula elucidation (HR2 formula tool).